from sklearn.datasets import make_classification
import pandas as pd
import numpy as np
import xgboost as xgb
from sklearn.model_selection import StratifiedGroupKFold, cross_val_score
import matplotlib.pyplot as plt2024-07-10
OBP:
We hebben vandaag de resultaten bekeken van de ranking (zie note 2024-06-30). Volgende denkrichtingen:
Train een XGBoost model dat een list van twee schedules als input neemt en als output de index van de schedule met de hoogste objective waarde. Zie: https://xgboost.readthedocs.io/en/latest/tutorials/learning_to_rank.html
Train een model dat in nauwkeurigheid toeneemt naarmate de objective value of ranking beter is. In dat geval zou de rankingplot meer conisch verlopen en de ranking nauwkeuriger zijn bij een betere ranking. Zie: https://elicit.com/notebook/aa0448de-dc7e-4d8b-8bd8-c1875679265f#17e0ddd35b0962672caf3894fb9da5b4
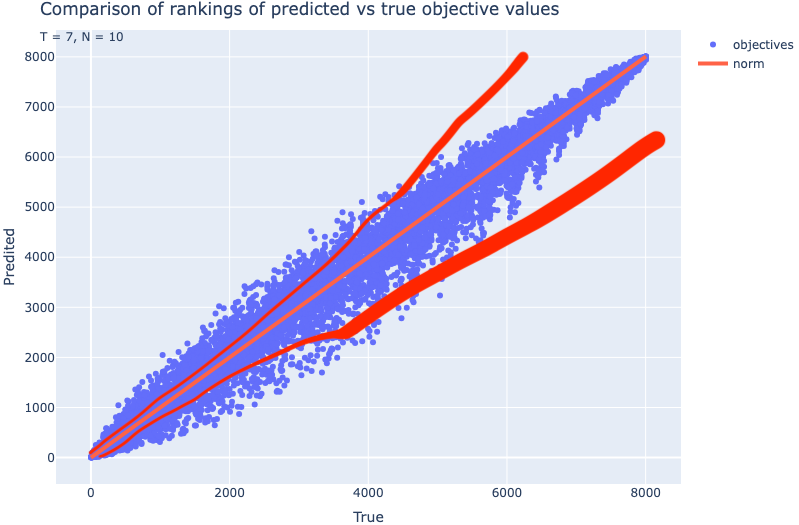
Ik test de snelheid van de het huidige XGBoost model t.o.v. de berekende waarde.
We maken een mix van berekening van de werkelijke waarde en berekening via het model. Hiervoor moeten we kijken of het model bij de waarde ook een inschatting van de betrouwbaarheid kan geven. Als de betrouwbaarheid laag is, wordt de werkelijke waarde berekend.
Mental note: Bekijk multiprocessing en joblib
Learning to rank
A synthetic classification dataset is an artificially generated dataset designed for classification tasks. These datasets are created programmatically rather than being collected from real-world observations. Synthetic datasets are often used for testing and demonstration purposes because they allow for controlled conditions and are readily available without the need for extensive data collection and preprocessing.
Purpose of Using Synthetic Datasets
Testing and Benchmarking: Synthetic datasets are useful for evaluating and comparing the performance of different machine learning algorithms under controlled conditions.
Algorithm Development: Researchers and developers can use synthetic data to develop and refine new algorithms without needing access to large real-world datasets.
Educational Use: Synthetic datasets are often used in educational settings to teach machine learning concepts and techniques.
Characteristics of Synthetic Classification Datasets
Controlled Parameters: You can specify various parameters such as the number of samples, number of features, number of informative and redundant features, class distribution, and noise level.
Predictability: Since the data is generated based on known parameters and distributions, the relationships between features and labels are well-defined. This can be useful for understanding how different algorithms perform under known conditions.
Reproducibility: By setting a random seed, synthetic datasets can be reproduced exactly. This is helpful for debugging and for comparing the performance of different models or algorithms under identical conditions.
Example of Synthetic Classification
In the provided code, the function make_classification from the sklearn.datasets module is used to generate a synthetic classification dataset:
Parameters of make_classification
The make_classification function allows you to specify a wide range of parameters to control the characteristics of the generated dataset. Here are some key parameters:
n_samples: The number of samples (data points) to generate.n_features: The total number of features in the dataset.n_informative: The number of informative features that are useful for predicting the target labels.n_redundant: The number of redundant features that are linear combinations of the informative features.n_classes: The number of distinct classes (labels).flip_y: The fraction of samples whose class labels are randomly flipped to introduce noise.class_sep: The factor that separates the classes, affecting the difficulty of the classification task.random_state: A seed for the random number generator to ensure reproducibility.
Workflow
The code generates a synthetic classification dataset and assigns each sample to one of three query groups. It then sorts the dataset based on the query group IDs. The purpose of this sorting is typically for tasks like ranking, where you need to organize the data by groups before applying certain algorithms or evaluations.
- Importing Libraries:
make_classificationfromsklearn.datasetsis used to generate a synthetic classification dataset.numpyis imported asnpfor numerical operations.xgboostis imported asxgb, though it is not used in the given code snippet.
Dataset Creation:
X: A 2D array where each row represents a sample and each column represents a feature.y: A 1D array of labels corresponding to each sample inX.
seed = 1994
X, y = make_classification(random_state=seed)- `seed` is set to 1994 to ensure reproducibility.
- `make_classification(random_state=seed)` generates a synthetic classification dataset `X` (features) and `y` (labels).Creating Query Groups:
rng: A random number generator seeded with1994to ensure the reproducibility of random operations.qid: A 1D array where each element is an integer (0, 1, or 2) indicating the query group ID for the corresponding sample inX.
rng = np.random.default_rng(seed)
n_query_groups = 3
qid = rng.integers(0, n_query_groups, size=X.shape[0])rngis initialized as a random number generator with the specifiedseed.n_query_groupsis set to 3, meaning we want to create 3 different query groups.qidis generated as an array of integers ranging from 0 ton_query_groups - 1(0, 1, 2) with the same length asX. Each integer represents the query group ID for the corresponding sample.
Sorting by Query Index:
sorted_idx: Indices that would sortqidin ascending order.- The arrays
X,y, andqidare then reordered usingsorted_idxso that samples belonging to the same query group are grouped together.
sorted_idx = np.argsort(qid)
X = X[sorted_idx, :]
y = y[sorted_idx]
qid = qid[sorted_idx]- Check data:
# Expand X into individual columns
X_expanded = pd.DataFrame(X, columns=[f'x_{i}' for i in range(X.shape[1])])
# Combine with qid and y
df = pd.concat([pd.DataFrame({'qid': qid}), X_expanded, pd.DataFrame({'y': y})], axis=1)
df.head()| qid | x_0 | x_1 | x_2 | x_3 | x_4 | x_5 | x_6 | x_7 | x_8 | ... | x_11 | x_12 | x_13 | x_14 | x_15 | x_16 | x_17 | x_18 | x_19 | y | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | -0.438941 | -0.518367 | -0.414652 | -1.031209 | -0.432312 | 1.252436 | 0.024844 | 0.539195 | 0.270635 | ... | -0.073392 | -0.855536 | -0.975249 | 1.240300 | 0.063219 | -1.498230 | 0.180950 | 1.129922 | -1.040233 | 1 |
| 1 | 0 | 0.198131 | -3.074477 | 0.933927 | 1.157325 | -2.341358 | 1.617548 | -0.687859 | 1.792938 | -0.541408 | ... | -1.000022 | -0.079594 | -0.828799 | -1.961206 | -0.821706 | 2.054504 | 0.250813 | -0.225271 | -0.120870 | 0 |
| 2 | 0 | -0.621530 | 0.732516 | 0.269019 | 0.559307 | 0.539464 | -1.104321 | 0.941721 | -1.897793 | 1.837516 | ... | 0.201258 | -0.109952 | 0.172811 | 0.331973 | 1.734853 | 0.154179 | -0.339768 | -0.495313 | 0.185678 | 1 |
| 3 | 0 | 0.388465 | 1.537994 | -2.087239 | 1.367650 | 0.377382 | 2.299039 | -1.719528 | -0.759369 | 1.483885 | ... | 0.783517 | -0.558472 | -2.253144 | 1.688623 | 1.633251 | -0.197652 | 0.589251 | -0.647990 | 2.221670 | 1 |
| 4 | 0 | -0.066125 | -0.926101 | 1.407887 | -0.773517 | 0.220194 | 0.474155 | 1.121773 | -1.250719 | -0.252777 | ... | 0.909320 | -0.399782 | -0.103039 | -0.908392 | 0.481251 | 0.081209 | 1.068396 | 0.725095 | -0.406591 | 0 |
5 rows × 22 columns
Initializing the ranker:
ranker = xgb.XGBRanker(...)initializes anXGBRankerobject with specific parameters:tree_method="hist": Specifies the tree construction algorithm to be used.lambdarank_num_pair_per_sample=8: Sets the number of pairs per sample for LambdaRank.objective="rank:ndcg": Sets the objective function to optimize Normalized Discounted Cumulative Gain (NDCG), which is common in ranking tasks.lambdarank_pair_method="topk": Specifies the pair generation method for LambdaRank.
Fitting the ranker:
ranker.fit(df, y)fits theXGBRankermodel to the data.dfis used as the feature matrix, andyis the target labels.Since
dfincludes theqidcolumn, the model can use this information to group samples correctly during training.
ranker = xgb.XGBRanker(tree_method="hist", lambdarank_num_pair_per_sample=8, objective="rank:ndcg", lambdarank_pair_method="topk")
## Exclude y from dataframe for the ranker
df = df.iloc[:, :-1]
# Check the distribution of query groups and labels
print("Query group distribution:", np.bincount(df['qid']))
print("Label distribution:", np.bincount(y))
ranker.fit(df, y)Query group distribution: [25 37 38]
Label distribution: [50 50]XGBRanker(base_score=None, booster=None, callbacks=None, colsample_bylevel=None,
colsample_bynode=None, colsample_bytree=None, device=None,
early_stopping_rounds=None, enable_categorical=False,
eval_metric=None, feature_types=None, gamma=None, grow_policy=None,
importance_type=None, interaction_constraints=None,
lambdarank_num_pair_per_sample=8, lambdarank_pair_method='topk',
learning_rate=None, max_bin=None, max_cat_threshold=None,
max_cat_to_onehot=None, max_delta_step=None, max_depth=None,
max_leaves=None, min_child_weight=None, missing=nan,
monotone_constraints=None, multi_strategy=None, n_estimators=None,
n_jobs=None, ...)In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
XGBRanker(base_score=None, booster=None, callbacks=None, colsample_bylevel=None,
colsample_bynode=None, colsample_bytree=None, device=None,
early_stopping_rounds=None, enable_categorical=False,
eval_metric=None, feature_types=None, gamma=None, grow_policy=None,
importance_type=None, interaction_constraints=None,
lambdarank_num_pair_per_sample=8, lambdarank_pair_method='topk',
learning_rate=None, max_bin=None, max_cat_threshold=None,
max_cat_to_onehot=None, max_delta_step=None, max_depth=None,
max_leaves=None, min_child_weight=None, missing=nan,
monotone_constraints=None, multi_strategy=None, n_estimators=None,
n_jobs=None, ...)fig, axes = plt.subplots(figsize = (20,20), dpi=500)
xgb.plot_tree(ranker, fontsize=14, ax=axes)
plt.show()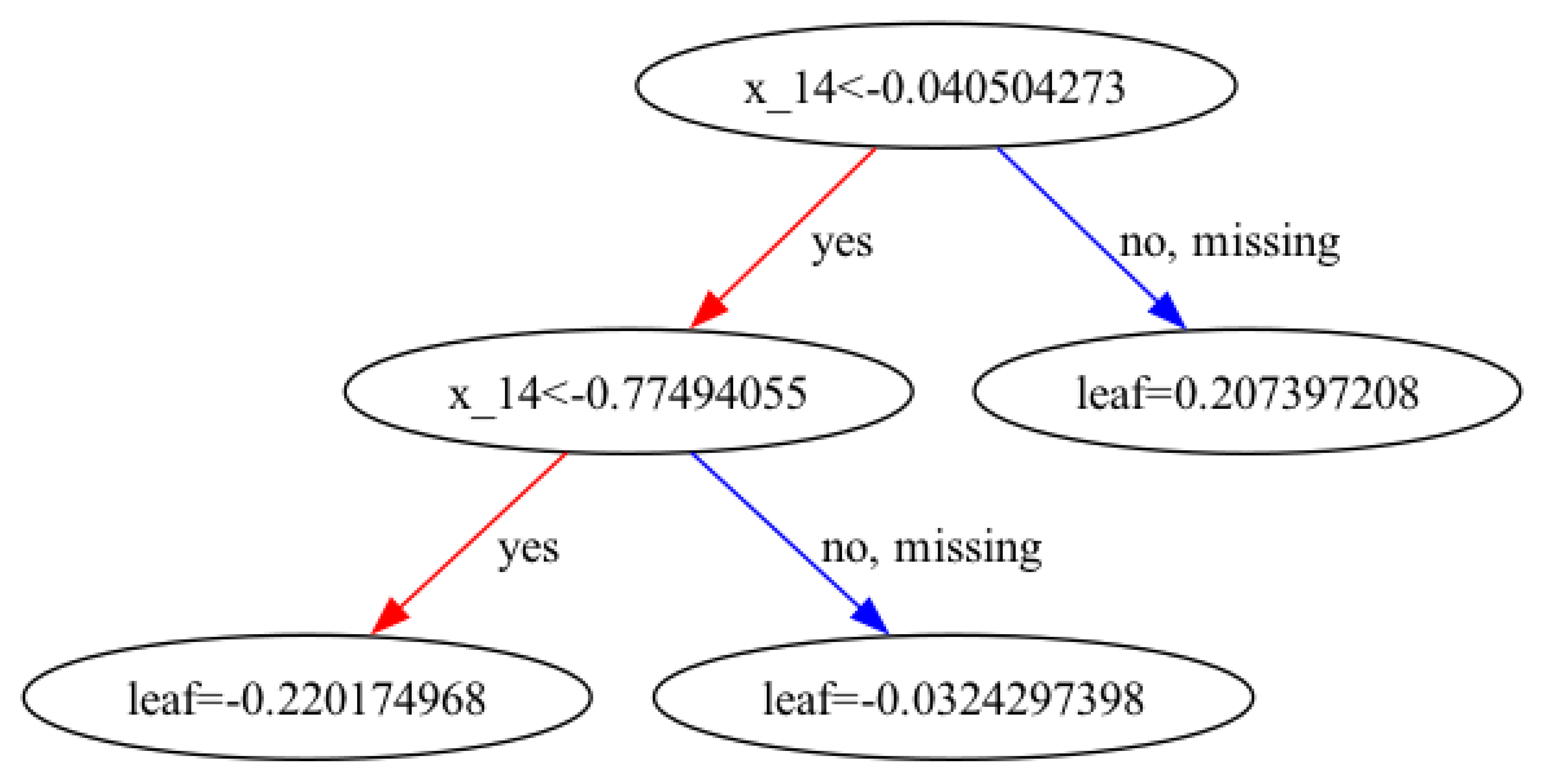
Cross-Validation:
kfold = StratifiedGroupKFold(shuffle=False)initializes aStratifiedGroupKFoldobject for cross-validation:shuffle=False: Specifies that the data should not be shuffled before splitting.StratifiedGroupKFoldensures that each fold maintains the same distribution of labels and respects the group structure (i.e., samples with the sameqidremain in the same fold).
cross_val_score(ranker, df, y, cv=kfold, groups=df.qid)performs cross-validation:ranker: TheXGBRankermodel to be evaluated.df: The feature matrix, including theqidcolumn.y: The target labels.cv=kfold: The cross-validation strategy.groups=df.qid: Ensures that samples with the same query ID (qid) are kept together in the same fold during cross-validation.
The function returns an array of scores from each fold.
# Works with cv in scikit-learn, along with HPO utilities like GridSearchCV
kfold = StratifiedGroupKFold(shuffle=False)
cross_val_score(ranker, df, y, cv=kfold, groups=df.qid)/Library/Frameworks/Python.framework/Versions/3.12/lib/python3.12/site-packages/xgboost/core.py:158: UserWarning:
[12:40:16] WARNING: /Users/runner/work/xgboost/xgboost/src/common/error_msg.cc:52: Empty dataset at worker: 0
array([1., 1., 1., 0., 0.])scores = ranker.predict(X)
sorted_idx = np.argsort(scores)[::-1]
# Sort the relevance scores from most relevant to least relevant
scores = scores[sorted_idx]
scoresarray([ 1.7109468 , 1.7109468 , 1.7109468 , 1.7109468 , 1.7109468 ,
1.7109468 , 1.7109468 , 1.7109468 , 1.7109468 , 1.7109468 ,
1.7109468 , 1.7109468 , 1.7109468 , 1.7109468 , 1.7109468 ,
1.7109468 , 1.7109468 , 1.7109468 , 1.7109468 , 1.7109468 ,
1.7109468 , 1.7109468 , 1.5921135 , 1.5921135 , 1.5921135 ,
1.5420871 , 1.5420871 , 1.5420871 , 1.5420871 , 1.5420871 ,
1.4591678 , 1.4591678 , 1.4232538 , 1.4232538 , 1.3153265 ,
1.3153265 , 0.97181636, 0.4532021 , -0.16912135, -0.4710336 ,
-0.4710336 , -1.0312457 , -1.0362681 , -1.0362681 , -1.2228703 ,
-1.2240134 , -1.2240134 , -1.2240134 , -1.2240134 , -1.2240134 ,
-1.2240134 , -1.2240134 , -1.3428468 , -1.3428468 , -1.3428468 ,
-1.4160769 , -1.4160769 , -1.4160769 , -1.4757924 , -1.4757924 ,
-1.4757924 , -1.6504426 , -1.6504426 , -1.6504426 , -1.6504426 ,
-1.6504426 , -1.655904 , -1.655904 , -1.655904 , -1.655904 ,
-1.655904 , -1.655904 , -1.655904 , -1.655904 , -1.655904 ,
-1.655904 , -1.655904 , -1.655904 , -1.655904 , -1.655904 ,
-1.655904 , -1.655904 , -1.8436491 , -1.8436491 , -1.8436491 ,
-1.8436491 , -1.8436491 , -1.8436491 , -1.8436491 , -1.8436491 ,
-1.8436491 , -1.8436491 , -1.8436491 , -1.8436491 , -1.8436491 ,
-1.8436491 , -1.8436491 , -1.8436491 , -1.8436491 , -1.8436491 ],
dtype=float32)